| Alias |
IND1, GT140, bHLH040, IND, EDA33 |
| TF Classification |
| FioreDB | bHLH (Click to show phylogenetic tree) |
| RARTF | bHLH / TRANSPARENT TESTA8 |
| AtTFDB | bHLH |
| PlnTFDB | bHLH (v3.0), bHLH (v1.0) |
| DATF | bHLH |
|
| TAIR short description |
INDEHISCENT, EMBRYO SAC DEVELOPMENT ARREST 33, basic helix-loop-helix (bHLH) DNA-binding superfamily protein (.1) |
| TAIR annotation |
INDEHISCENT (IND); FUNCTIONS IN: DNA binding, sequence-specific DNA binding transcription factor activity; INVOLVED IN: polar nucleus fusion, regulation of transcription; LOCATED IN: nucleus; CONTAINS InterPro DOMAIN/s: Helix-loop-helix DNA-binding domain (InterPro:IPR001092), Helix-loop-helix DNA-binding (InterPro:IPR011598); BEST Arabidopsis thaliana protein match is: basic helix-loop-helix (bHLH) DNA-binding superfamily protein (TAIR:AT5G09750.1); Has 2280 Blast hits to 2276 proteins in 91 species: Archae - 0; Bacteria - 0; Metazoa - 0; Fungi - 0; Plants - 2280; Viruses - 0; Other Eukaryotes - 0 (source: NCBI BLink). (.1) |
| Gene model |
|
| Entry clone (w/o stop) |
IE_01D04 |
| External link |
|
| Gene Ontology (GO) |
| (P) 10197: cell organization and biogenesis |
->polar nucleus fusion |
| (F) 3677: DNA or RNA binding |
->DNA binding |
| (F) 3700: transcription factor activity |
->transcription factor activity, sequence-specific DNA binding |
| (F) 46983: protein binding |
->protein dimerization activity |
| (C) 5634: nucleus |
->nucleus |
| (P) 6351: transcription,DNA-dependent |
->transcription, DNA-templated |
| (P) 6355: other metabolic processes |
->regulation of transcription, DNA-templated |
|
| miRNA/tasiRNA |
|
| Orthologs |
|
Type |
| Ia |
Ib |
II |
III |
Total |
| poplar |
|
|
|
|
|
| soybean |
|
|
|
|
|
| rice |
|
|
|
|
|
| sorghum |
|
|
|
|
|
| physcomitrella |
|
|
|
|
|
| Total |
|
|
|
|
0E0 |
|
| Repression motif |
|
Repression motif
in putative orthologs |
|
| Status |
| Plasmid construction | -> done |
| Plasmid construction | -> done |
| Transformed | -> done |
| T1 seed harvested | -> done |
| T2 seed harvested | -> done |
|
CRES-T phenotype from individual project
View all photo |
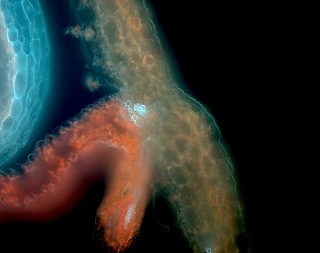
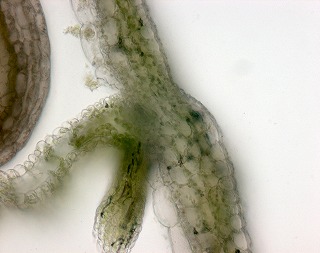
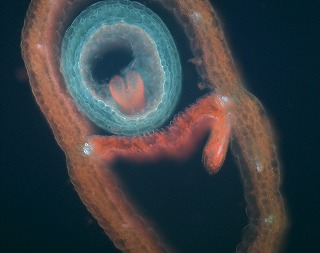
Upward curling rosette leaf very similar to NST1 over-expression, but no ectopic secondary wall was observed. Differentiation of valve margin is suppressed and lignification of endodrmal layer of valve is also suppressed, but not practical due to very low fertility.
abnormal silique (direct input),
short pedicel (direct input),
hyponastic leaf (direct input),
abnormal leaf (direct input),
sterile or low fertility (direct input)
      and more... and more... |
| CRES-T phenotype from publication |
|
| CRES-T phenotype from bulk project |
rough surface leaf,
hyponastic leaf
(A20355-180)


rough surface leaf,
hyponastic leaf
(A20354-123)


|
Phenotype in ornamental plants
|
|
| Comment |
|
