| Alias |
ATMYBRTF, ATMYB23 |
| TF Classification |
|
| TAIR short description |
myb domain protein 23 (.1) |
| TAIR annotation |
Encodes a MYB gene that, when overexpressed ectopically, can induce ectopic trichome formation. It is a member of subgroup 15, together with WER and GL1. Members of this subgroup share a conserved motif of 19 amino acids in the putative transcription activation domain at the C-terminal end. The gene is expressed in leaves, stems, flowers, seeds and roots and quite strongly in trichomes. There is partial functional redundancy between ATMYB23 and GL1. The two proteins are functionally equivalent with respect to the regulation of trichome initiation but not with respect to trichome branching - which is controlled by MYB23 and not GL1. (.1) |
| Gene model |
|
| Entry clone (w/o stop) |
RE_02H10 |
| External link |
|
| Gene Ontology (GO) |
| (P) 10026: other cellular processes |
->trichome differentiation |
| (P) 10053: other cellular processes |
->root epidermal cell differentiation |
| (P) 10091: cell organization and biogenesis |
->trichome branching |
| (F) 1135: other molecular functions |
->transcription factor activity, RNA polymerase II transcription factor recruiting |
| (F) 3677: DNA or RNA binding |
->DNA binding |
| (F) 3700: transcription factor activity |
->transcription factor activity, sequence-specific DNA binding |
| (F) 43565: DNA or RNA binding |
->sequence-specific DNA binding |
| (F) 44212: DNA or RNA binding |
->transcription regulatory region DNA binding |
| (P) 48629: other cellular processes |
->trichome patterning |
| (F) 5515: protein binding |
->protein binding |
| (C) 5634: nucleus |
->nucleus |
| (P) 6351: transcription,DNA-dependent |
->transcription, DNA-templated |
| (P) 6355: transcription,DNA-dependent |
->regulation of transcription, DNA-templated |
| (P) 6357: other metabolic processes |
->regulation of transcription from RNA polymerase II promoter |
| (F) 981: transcription factor activity |
->RNA polymerase II transcription factor activity, sequence-specific DNA binding |
|
| miRNA/tasiRNA |
|
| Orthologs |
|
Type |
| Ia |
Ib |
II |
III |
Total |
| poplar |
|
|
1 |
1 |
2 |
| soybean |
|
|
|
|
|
| rice |
|
|
|
|
|
| sorghum |
|
|
|
|
|
| physcomitrella |
|
|
|
|
|
| Total |
|
|
1 |
1 |
2 |
|
| Repression motif |
|
Repression motif
in putative orthologs |
|
| Status |
| Plasmid construction | -> done |
| Plasmid construction | -> done |
| Transformed | -> done |
| T1 seed harvested | -> done |
| T2 seed harvested | -> done |
|
CRES-T phenotype from individual project
View all photo |
hairy root formation, elongated leaves and stems, defectin the deposition of mucilage on seed coat
glabra (direct input),
abnormal stem (direct input),
too large leaf (direct input),
too many roothairs (direct input),
abnormal seed (direct input)
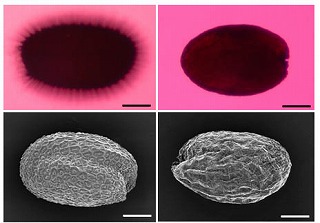  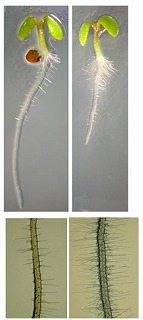 |
| CRES-T phenotype from publication |
glabra, too many roothairs, abnormal seed (PMID15668208)
|
| CRES-T phenotype from bulk project |
|
Phenotype in ornamental plants
|
| ID |
Species |
Construct |
Date |
Comment |
Phenotype |
Photo |
| ID50010 |
Torenia |
35S:AtCDS-SRDX |
2005-10-24 |
|
no phenotype |
|
| ID50009 |
Chrysanthemum |
35S:AtCDS-SRDX |
2005-10-24 |
|
no phenotype |
|
| ID59 |
Cyclamen |
35S:AtCDS-SRDX |
2005-09-08 |
|
no phenotype |
|
|
| Comment |
|
