| Alias |
OBP2, AtDof1. 1 |
| TF Classification |
|
| TAIR short description |
Dof-type zinc finger DNA-binding family protein (.1.2.3) |
| TAIR annotation |
A member of the DOF transcription factors. Prominently expressed in the phloem of leaves and other organs. Expression is induced by wounding, MeJA and insect feeding. Upregulates glucosinolate biosynthesis. (.1.2.3) |
| Gene model |
|
| Entry clone (w/o stop) |
RE_0465 |
| External link |
|
| Gene Ontology (GO) |
| (P) 10439: other metabolic processes |
->regulation of glucosinolate biosynthetic process |
| (P) 3002: developmental processes |
->regionalization |
| (F) 3677: DNA or RNA binding |
->DNA binding |
| (F) 3700: transcription factor activity |
->transcription factor activity, sequence-specific DNA binding |
| (F) 44212: DNA or RNA binding |
->transcription regulatory region DNA binding |
| (F) 46872: other binding |
->metal ion binding |
| (C) 5634: nucleus |
->nucleus |
| (P) 6351: transcription,DNA-dependent |
->transcription, DNA-templated |
| (P) 6355: other cellular processes |
->regulation of transcription, DNA-templated |
| (P) 9611: response to stress |
->response to wounding |
| (P) 9625: response to abiotic or biotic stimulus |
->response to insect |
| (P) 9753: other biological processes |
->response to jasmonic acid |
|
| miRNA/tasiRNA |
|
| Orthologs |
|
Type |
| Ia |
Ib |
II |
III |
Total |
| poplar |
|
|
|
|
|
| soybean |
|
|
|
|
|
| rice |
|
|
|
|
|
| sorghum |
|
|
|
|
|
| physcomitrella |
|
|
|
|
|
| Total |
|
|
|
|
0E0 |
|
| Repression motif |
|
Repression motif
in putative orthologs |
|
| Status |
| Plasmid construction | -> done |
| Plasmid construction | -> done |
| Transformed | -> done |
| T1 seed harvested | -> done |
| T2 seed harvested | -> done |
|
| CRES-T phenotype from individual project
|
stem elongation in the vegetative phase, fusion of stem and petiole, early flowering?, cotyledon morphology, lack of shoot meristem
fused stem (direct input),
meristem-less (direct input),
early flowering (direct input)
|
| CRES-T phenotype from publication |
|
| CRES-T phenotype from bulk project |
loss of apical dominance,
short pedicel,
pollen defective,
abnormal carpel,
narrow leaf,
too many rosettes,
too few branching,
sterile or low fertility
(A20258-148)

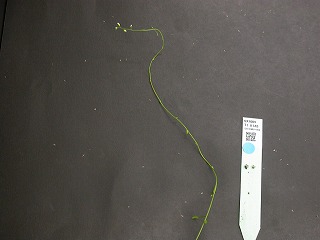


narrow leaf,
abnormal phyllotaxis,
too many rosettes
(A20585-153)
aerial rosette,
pollen defective,
abnormal carpel,
sterile or low fertility
(A20258-113)



|
Phenotype in ornamental plants
|
|
| Comment |
|
